Tagmentation input calculation:¶
Note: During the Tagmentation process additional sequencing adapters are added and the library size is further reduced for Illumina Sequencers. Tagmentation is sensitive to the input DNA concentraion and the average length of DNA fragments used. Therefore to get consistent Post-tagmentation average library sizes between 570-620bp we developed an equation to help you calculate the input DNA amount.
For Estimated Center in bp<=1350: $$ Input\ DN A\ in\ ng = 1.37692 - 0.000576923\times{Estimated\ Center\ in\ bp} $$
For Estimated Center in bp >1350 we always use 0.6ng as input.
You will need the Tapestation HS D5000 plot to estimate the center of the input DNA fragment distribution. Each Tapestation Plot comes with Labels on the X axis which tells you the number of bases for each part of the DNA Fragment Distribution. Generally the labels of the peaks coincide with the actual center of the DNA fragment distribution. But sometimes they don't. In case they don't you will have to visually estimate the center by looking at the DNA Fragment distribution. Ignore the Upper and Lower Markers when trying to estimate the Center. The following examples should help you estimate the center in bp of your DNA fragment distribution.
TO DO: Use better example Tapestations for this section. Use plots with no primer peak.
| Example 1 | Example 2 |
|---|---|
| Estimated Center: 986bp Calculated Input DNA in ng: 0.81 |
Estimated Center: 968bp Calculated Input DNA in ng: 0.82 |
 |
 |
| Tagmented Library Center: 593bp | Tagmented Library Center: 620bp |
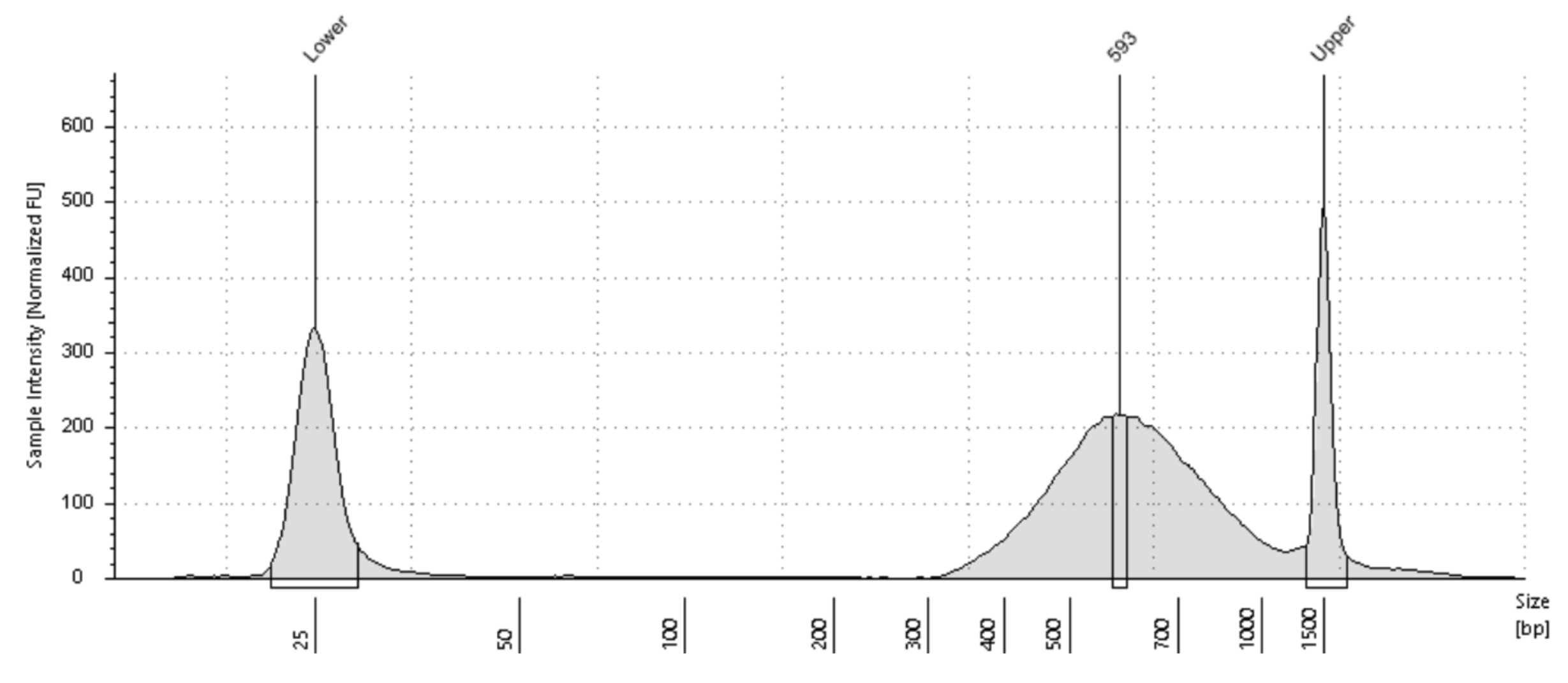 |
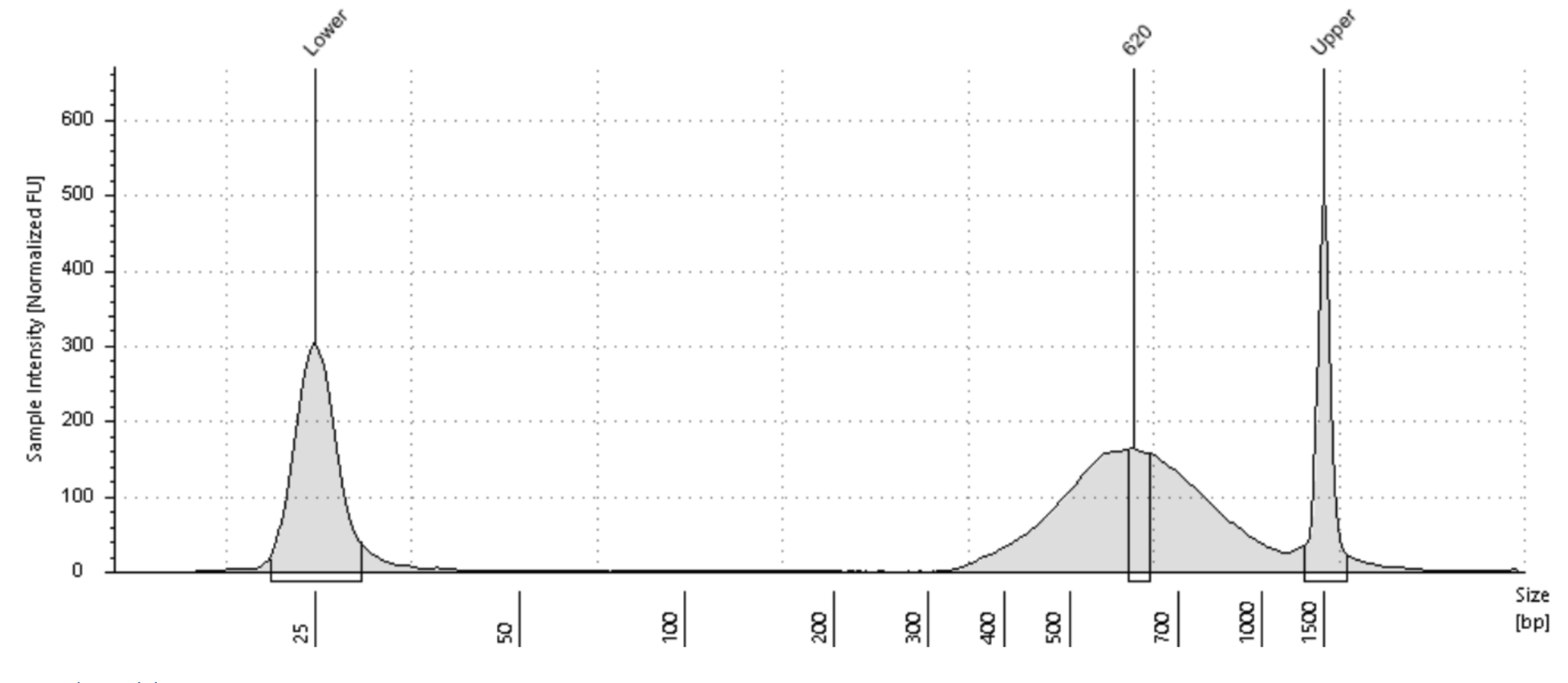 |